Let me start off by stating that I have enormous respect for Rob Edgar (creator of USEARCH, UPARSE etc.). His contributions to the field of bioinformatics, and indirectly to the fields of molecular and microbial ecology have been huge, you only need to look at his citation rates to see that! So this post is not intended as a criticism of him or his work in any way.
That said, I’ve recently been thinking about the deluge of new algorithms for picking Operational Taxonomic Units (OTUs) from molecular sequence datasets and wondering where, and how, USEARCH, UCLUST et al. will fit in.
Rob Edgar only made 32-bit versions of all his software freely available for users, with users having to pay $885 for a 64-bit license. A 32-bit license essentially limits the amount of RAM your computer can access whilst using this software to 4gb. So even if you had a cluster computer at your disposal, you’d still be shackled by the 4gb limit. A few years ago, when 454 pyrosequencing was the predominant sequencing platform, this wasn’t so much of a problem because the datasets were big, but not huge. However, with the rise of Illumina’s platforms such as the HiSeq and MiSeq, molecular datasets are becoming truly enormous, meaning more RAM is needed to work with them.
Furthermore, the arrival of VSEARCH, a 64-bit open source work-alike of USEARCH, has meant that you now don’t need to shell out for an expensive 64-bit license to get USEARCH-esque results. This leads me to wonder, what is the future of USEARCH now that there are good, free, and open-source alternatives?
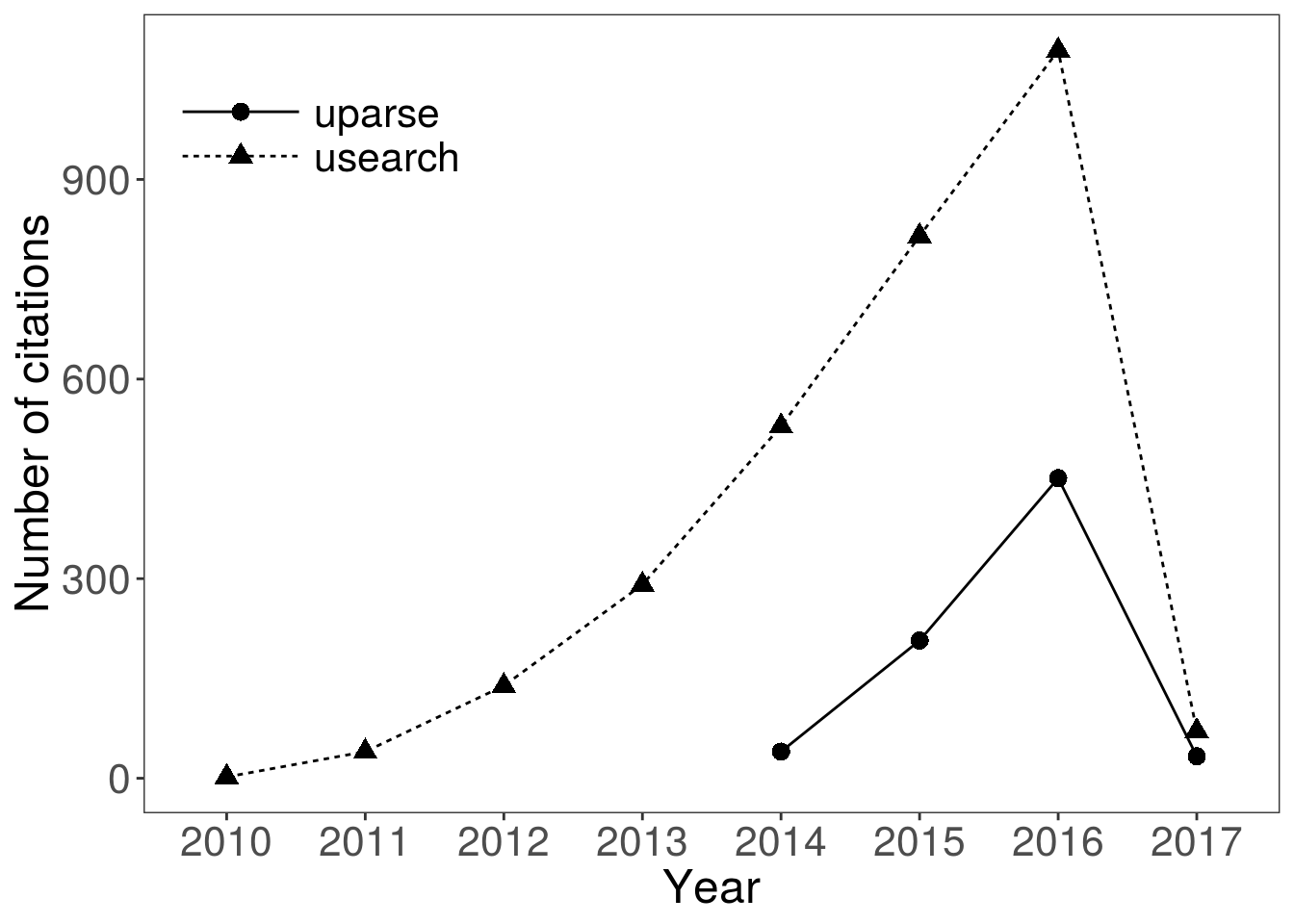
Looking at the citation rates of Edgar’s papers, there are no obvious signs of it losing popularity… Data on the plot below were downloaded using Web of Science. It will be interesting to see if the popularity of this classic piece of bioinformatics software ever declines.